Second-generation Amine Reactive Biosensor Advances the Octet Platform
Wesley McGinn-Straub, Product Manager, ForteBio
Amine coupling using EDC and NHS or sulfo-NHS is a classic method of coupling a carboxylate to a primary amine to form a covalent amide bond. The chemistry has been widely used in label-free assays to immobilize proteins and other biomolecules on biosensors for kinetic analysis of molecular interactions. The method is general, rapid, convenient and irreversible. The entire reaction is typically executed within 10 to 15 minutes. It requires no prior preparation and forms a bond stable through a wide range of temperature and pH.
Buoyed by the experience of developing 13 biosensors, ForteBio recently took aim at enhancing the performance of its original Amine Reactive (AR) biosensor in order to expand the applications that can be performed using amine coupling on the Octet platform. The effort produced a significant advancement in biosensor chemistry, resulting in the introduction of the Amine Reactive 2nd Generation (AR2G) biosensor for kinetics.
The AR2G biosensor chemistry is entirely new. Every reagent layer, from the silica surface of the fiber optic to the final carboxylates used for coupling, has been re-engineered to produce a surface with multiple assay benefits, including increased loading density, increased concentration of functional ligand and increased immobilization pH robustness. The AR2G biosensor therefore offers potential success in assays unapproachable using the AR biosensor; for example, analytes under 20 kD or binding interactions with weaker affinity constants.
The newly engineered AR2G biosensor contains a higher concentration of carboxylates than the AR biosensor, enabling a higher ligand loading density. A comparison of the loading steps for a library of protein ligands demonstrated that the AR2G biosensors achieved an average 3-fold greater loading density than the AR biosensors. Four examples of the increased loading density of the AR2G biosensors versus that of the AR biosensors are shown in Figure 1.
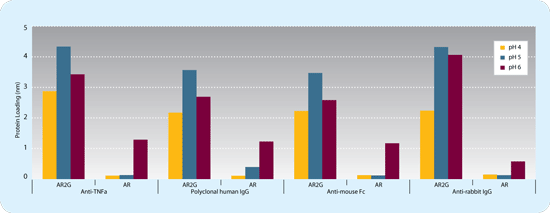
Figure 1: Performance comparison of AR2G and AR biosensors. Four proteins (20 µg/mL) were immobilized on AR2G biosensors using EDC/sulfo-NHS (acetate buffer at pH 4, 5 and 6) and on AR biosensors using EDC-NHS (MES buffer at pH 4, 5 and 6). The signal intensity achieved with the AR2G biosensors during loading was significantly greater than that with the AR biosensors for all proteins. The average increase was approximately four-fold with a maximum increase of 7.5-fold (anti-rabbit IgG). The enhanced performance of the AR2G biosensor was robust towards pH with increased loading density observed at all pH values.
Increased ligand loading density on the AR2G biosensor also translates to an average 3.5-fold signal enhancement compared to AR biosensor. Examples were also observed in which the AR2G biosensors generated greater signal amplitude during analyte detection than the AR biosensors, despite equal ligand loading on the two biosensors. The AR2G biosensor therefore enables not only more immobilized ligand but more functional immobilized ligand. The increased sensitivity enables detection of smaller analytes, lower affinity analytes and analytes of lower concentration, important during kinetic titrations where low ligand loading densities and low analyte concentrations often produce the best conditions for monitoring binding kinetics.
The AR2G biosensor also provides a new alternative to the Streptavidin (SA) biosensor. While the Streptavidin biosensor still has a higher loading capacity than the AR2G biosensor due to the heterogeneous nature of proteins, kinetic analyses of some systems have been shown to be more successful with AR2G biosensors than SA biosensors. Due to the highly heterogeneous properties of proteins and other molecules, performance of the AR2G biosensor will vary for each binding pair.
Amine coupling chemistry is generally performed at an acidic pH between 4.0 and 6.0 to create a slight positive charge on the protein, which promotes association with the negatively charged carboxylate surface on the biosensor, which in turn faciliatates the immobilization event. Typically, the optimal pH for immobilization is determined by scouting the assay at pH 4, 5 and 6. The new AR2G biosensor demonstrates a dramatic robustness in immobilization pH, largely eliminating the need for a pH scout for most assays. Approximately 9 out of 10 proteins evaluated for using the AR2G biosensor had an optimum immobilization pH of 5. For the remaining proteins, the values achieved with pH 5 were within 85% of pH 4 and pH 6, indicating good signal strength is likely to be achieved in this pH range. For most protein systems, therefore, the ligand can be immobilized at pH 5 and then directly assayed for analyte binding, saving time and improving throughput. Note that for assays with very low signal or difficult to detect analytes (either due to molecular weight or low affinity), a pH scout is recommended.
Finally, many of the newest and most interesting proteins are available in limited supply — with a large number of analytical assays prioritized to answer the early and essential questions in drug discovery. For those precious samples, the synergy of the user-customizable AR2G biosensor and the microfluidics-free, microplate-based Octet platform offer a unique analytical advantage. Using AR2G biosensors, a custom kinetic biosensor can be prepared from a protein (or any other primary amine containing molecule) with little or no sample consumption. Because the instrument uses a microplate format, free of microfluidics, no sample is lost due to injection cycles and there is no minimum aliquot required for processing steps such as biotinylation.
For more information on the AR2G biosensor and its potential use in your research efforts, please contact your local ForteBio representative or download the AR2G biosensor datasheet and technical notes from the ForteBio website. |